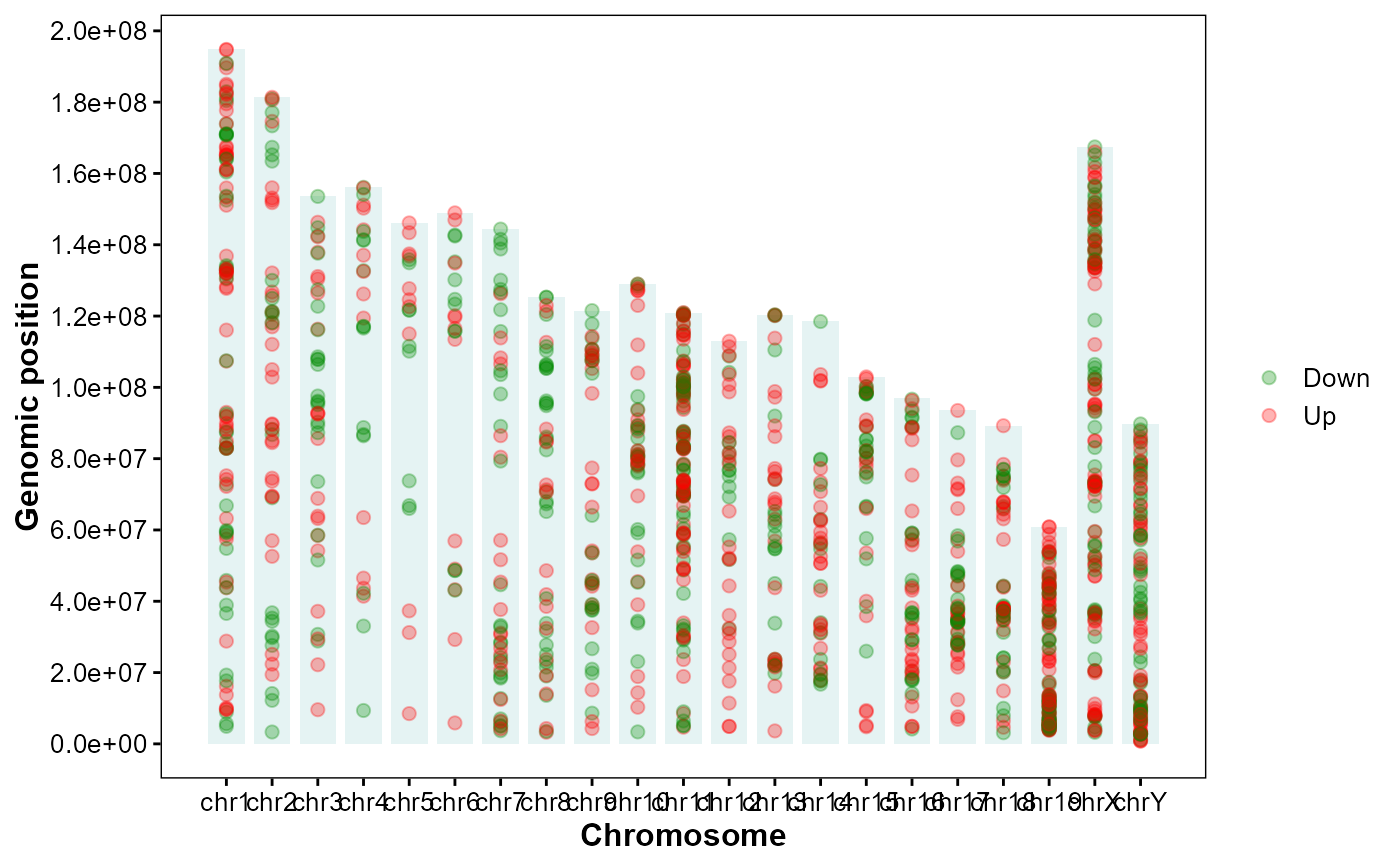
Plot DEGs up/down along chromosomes.
Usage
plot_deg_exp(
deg_file,
gff_file,
format = "auto",
id_col = "GeneID",
fc_col = "log2FoldChange",
orientation = "horizontal",
chrom_alpha = 0.1,
chrom_color = "#008888",
bar_height = 0.8,
point_size = 2,
point_alpha = 0.3,
up_color = "#ff0000",
down_color = "#008800",
mark_style = "point",
line_width = 0.6,
line_height = 0.8
)Arguments
- deg_file
DEG table from DESeq2 analysis.
- gff_file
Genomic structural annotation
GFF3/GTFfile path.- format
Format of GFF3/GTF file. ("auto", "gff3", "gtf").
- id_col
Gene IDs column name. ("GeneID").
- fc_col
Log2(fold change) column name. ("log2FoldChange").
- orientation
Coordinate orientation. ("horizontal", "vertical").
- chrom_alpha
Chromosome bar alpha. (0.1).
- chrom_color
Chromosome bar color. ("#008888").
- bar_height
Chromosome bar thickness in y units. (0.8).
- point_size
Point size. (2).
- point_alpha
Point alpha. (0.3).
- up_color
Color for up-regulated genes. ("#ff0000").
- down_color
Color for down-regulated genes. ("#008800").
- mark_style
Marker style for DEGs. ("point", "line").
- line_width
Line width. (0.6).
- line_height
Line height relative to bar radius. (0.8).
Examples
# DEG results from DESeq2
deg_file <- system.file(
"extdata",
"example.deg",
package = "GAnnoViz")
# Genomic structure annotation
gff_file <- system.file(
"extdata",
"example.gff3.gz",
package = "GAnnoViz")
# Plot DEGs along chromosomes
plot_deg_exp(
deg_file = deg_file,
gff_file = gff_file,
format = "auto",
id_col = "GeneID",
fc_col = "log2FoldChange",
orientation = "horizontal",
chrom_alpha = 0.1,
chrom_color = "#008888",
bar_height = 0.8,
point_size = 2,
point_alpha = 0.3,
up_color = "#ff0000",
down_color = "#008800",
mark_style = "point",
line_width = 0.6,
line_height = 0.8)
#> Import genomic features from the file as a GRanges object ...
#> OK
#> Prepare the 'metadata' data frame ...
#> OK
#> Make the TxDb object ...
#> OK