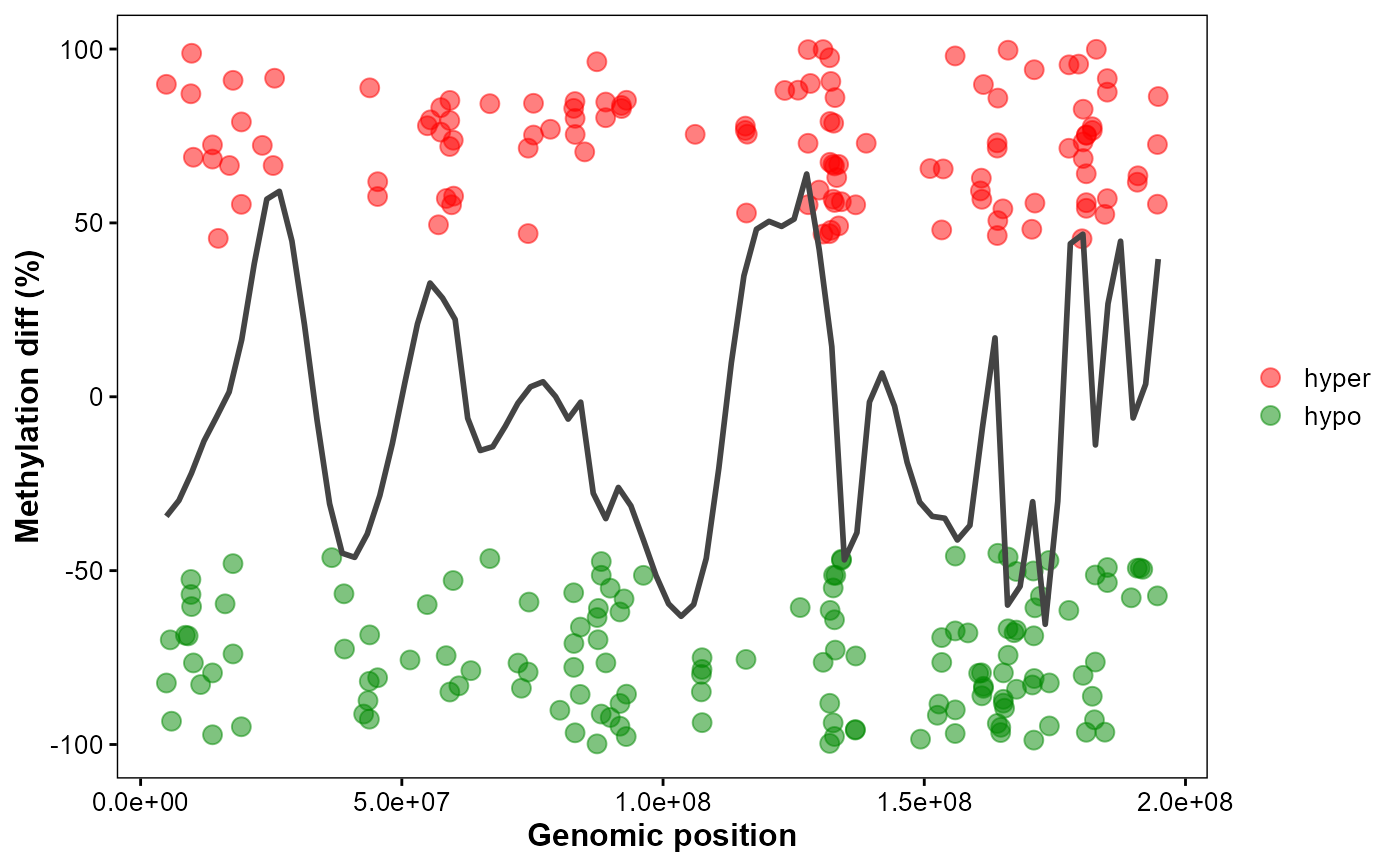
Plot chromosomal DMGs trend.
Usage
plot_dmg_trend(
chrom_id,
dmr_file,
smooth_span = 0.1,
hyper_color = "#ff000055",
hypo_color = "#00880055",
point_size = 3,
point_alpha = 0.5
)Arguments
- chrom_id
Chromosome identifier (e.g., "chr1").
- dmr_file
DEG table from MethylKit analysis.
- smooth_span
Span for local regression smoothing. (0.1).
- hyper_color
Color for hyper-methylated DMRs. ("#ff0000").
- hypo_color
Color for hypo-methylated DMRs. ("#008800").
- point_size
Point size. (3).
- point_alpha
Point alpha. (0.5).
Examples
# Example DMR table in GAnnoViz
dmr_file <- system.file(
"extdata",
"example.dmr",
package = "GAnnoViz")
# Plot DMR trend
plot_dmg_trend(
chrom_id = "chr1",
dmr_file = dmr_file,
smooth_span = 0.1,
hyper_color = "#ff0000",
hypo_color = "#008800",
point_size = 3,
point_alpha = 0.5
)
#> `geom_smooth()` using method = 'loess' and formula = 'y ~ x'