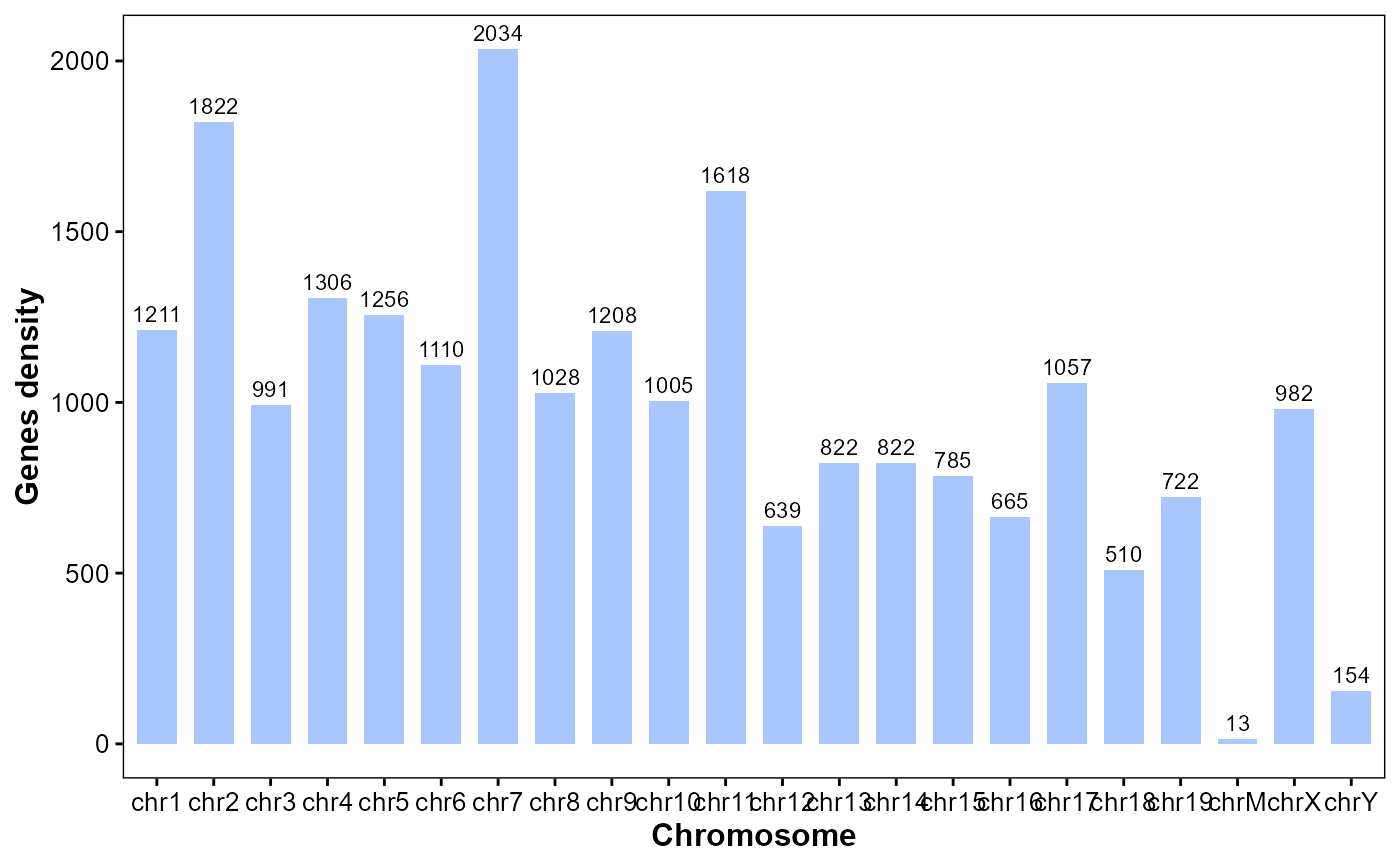
Plot gene stats for chromosomes.
Usage
plot_gene_stats(
gff_file,
format = "auto",
bar_width = 0.7,
bar_color = "#0055ff55",
lable_size = 3
)Examples
# Example GFF3 file in GAnnoViz
gff_file <- system.file(
"extdata",
"example.gff3.gz",
package = "GAnnoViz")
# Plot gene stats
plot_gene_stats(
gff_file = gff_file,
format = "auto",
bar_width = 0.7,
bar_color = "#0055ff55",
lable_size = 3)
#> Import genomic features from the file as a GRanges object ...
#> OK
#> Prepare the 'metadata' data frame ...
#> OK
#> Make the TxDb object ...
#> OK